A Python Utility for Wrapping Rosetta Macromolecural Modeling Suite.
Caution
RosettaPy requires Rosetta compiled and installed.
Before running RosettaPy, please DO make sure that you have obtained the correct license from Rosetta Commons.
For more details, please see this page.
Important
RosettaPy is NOT PyRosetta.
You probably don't need to install this package if you are looking for PyRosetta.
Please see this page.
RosettaPy is designed to handle the complexities of locating, composing and running Rosetta binaries within Python.
The module includes:
| Key Component | Description |
|---|---|
| Rosetta | A command-line wrapper for Rosetta that simplifies the setting up and running commands. |
| RosettaScriptsVariableGroup | Represents variables used in Rosetta scripts, facilitating their management and use. |
| RosettaEnergyUnitAnalyser | Analyzes and interprets Rosetta output score files, providing a simplified interface for result analysis. |
| Example Applications | Demos of specific Rosetta applications like PROSS, FastRelax, RosettaLigand, Supercharge, MutateRelax, and Cartesian ddG for different computational biology tasks. |
One can install RosettaPy directly from PyPI:
pip install RosettaPy -U-
Import necessary modules
from RosettaPy import Rosetta, RosettaScriptsVariableGroup, RosettaEnergyUnitAnalyser from RosettaPy.node import RosettaContainer, MpiNode, Native
-
Create a
Rosettaproxy with parametersrosetta = Rosetta( # a binary name for locating the real binary path bin="rosetta_scripts", # flag file paths (please do not use `@` prefix here) flags=[...], # command-line options opts=[ "-in:file:s", os.path.abspath(pdb), "-parser:protocol", "/path/to/my_rosetta_scripts.xml", ], # output directory output_dir=..., # save pdb and scorefile together save_all_together=True, # a job identifier job_id=..., # silent the rosetta logs from stdout verbose = False, )
RosettaPy uses
Nativenode by default.It has to be noted that
NativeandMpiNodeare only available on Linux and macOS. For Windows users, please refer to theFull Operating System Compatibility TableandGetting Windows Ready for Rosetta Runssections below. -
Compose rosetta tasks matrix as inputs
tasks: list[Dict[st 8000 r, Any]] = [ # Create tasks for each variant { "rsv": RosettaScriptsVariableGroup.from_dict( { "var1": ..., "var2": ..., "var3": ..., } ), "-out:file:scorefile": f"{variant}.sc", "-out:prefix": f"{variant}.", } for variant in variants ] # pass task matrix to rosetta.run as `inputs` rosetta.run(inputs=tasks)
-
Using structure labels (
-nstruct)Create distributed runs with structure labels (
-nstruct <int>) is feasible. For local runs without MPI or container,RosettaPyimplemented this feature by ignoring the build-in job distributer of Rosetta, canceling the default output structure label, attaching external structural label as unique job identifier to each other, then run these tasks only once for each. This enables massive parallalism.options=[...] # Passing an optional list of options that will be used to all structure models rosetta.run(nstruct=nstruct, inputs=options) # input options will be passed to all runs equally
-
Call Analyzer to check the results
analyser = RosettaEnergyUnitAnalyser(score_file=rosetta.output_scorefile_dir) best_hit = analyser.best_decoy pdb_path = os.path.join(rosetta.output_pdb_dir, f'{best_hit["decoy"]}.pdb') # Ta-da !!! print("Analysis of the best decoy:") print("-" * 79) print(analyser.df.sort_values(by=analyser.score_term)) print("-" * 79) print(f'Best Hit on this run: {best_hit["decoy"]} - {best_hit["score"]}: {pdb_path}')
One can also build a customized analyser by re-using the
analyser.dfDataframe.
Here are some tips for advanced usages to adjust the workflow in respect to behaviors of some Rosetta workflows and applications.
Some Rosetta Apps (Superchange, Cartesian ddG, etc.) may produce files at their working directory, and this may not threadsafe if one runs multiple jobs in parallel in the same directory. In this case, the isolation flag can be used to create a temporary directory for each run.
Rosetta(
...
+ isolation=True,
)There are various node configurations that can be used to run Rosetta jobs.
| Node | Linux | macOS | Windows | Architectures | Prerequisite |
|---|---|---|---|---|---|
| Native | ✅1 | ✅ | ❌ | x86_64, aarch64 | Rosetta compiled. |
| MpiNode | ✅1 | ✅ | ❌ | x86_64, aarch64 | Rosetta compiled with extras=mpi flag; MPI installed |
| RosettaContainer | ✅ | ✅2 | ✅ | x86_64 | Docker or Docker Desktop installed and launched. |
| WslWrapper3 | ❌ | ❌ | ✅1 | x86_64, aarch644 | WSL2, with Rosetta built and installed on. |
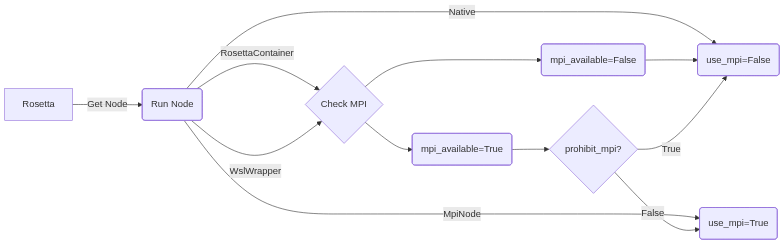
This Flow chart shows how RosettaPy decides whether to use MPI or not.
Before RosettaContainer or WslWrapper instance is passed as run_node, the node itself checks if MPI-related environment is ready. Also, the flag prohibit_mpi will force to disable MPI if it is True.
By default, RosettaPy uses Native node with 4 CPU cores, representing the local machine with Rosetta installed.
To specify the number of cores, use the nproc parameter.
Rosetta(
...
+ run_node=Native(nproc=8)
)Requires MPI(mpich, openmpi, etc.) installed.
If one wish to run with Rosetta that was installed on local and built with extra=mpi flag via MPI,
consider using MpiNode instance as run_node instead. This enables native parallelism feature with MPI.
Rosetta(
...
+ run_node=MpiNode(nproc=10),
)Also, if one wishes to use MpiNode with Slurm task manager, specifying run_node to MpiNode.from_slurm() may help
with fetching the node info from the environment.
This is an experimental feature that has not been seriously tested in production.
Rosetta(
...
+ run_node=MpiNode.from_slurm(),
)If one wishes to use the Rosetta container as the task worker, (WSL + Docker Desktop, for example)
setting a run_node option as RosettaContainer class would tell the proxy to use it.
This image names can be found at https://hub.docker.com/r/rosettacommons/rosetta
Note that the paths of each task will be mounted into the container and rewritten to the container's path.
This rewriting feature may fail if the path is mixed with complicated expressions as options.
Non-mpi image:
Rosetta(
...
+ run_node=RosettaContainer(image="rosettacommons/rosetta:latest"),
)or MPI image:
Rosetta(
...
+ run_node=RosettaContainer(image="rosettacommons/rosetta:mpi"),
+ use_mpi=True, # one still needs to enable MPI by this flag if MPI is required.
)or non-MPI runs with MPI image:
Rosetta(
...
+ run_node=RosettaContainer(image="rosettacommons/rosetta:mpi", prohibit_mpi=True), # override MPI setting
)During the workflow processing, one will see some active containers at Containers tab of Docker Desktop, if Docker Desktop is installed. Also, typing docker ps in the terminal will show them too. Each of these containers will be destructed immediately after its task finished or stopped.
One can still pick the desire node quickly by calling node_picker method.
from RosettaPy.node import node_picker, NodeHintT
node_hint: NodeHintT = 'docker_mpi'
Rosetta(
...
+ run_node=node_picker(node_type=node_hint),
+ use_mpi=...,
)Where node_hint is one of ["docker", "docker_mpi", "mpi", "wsl", "wsl_mpi", "native"]
Caution
AGAIN, before using this tool, please DO make sure that you have licensed by Rosetta Commons. For more details of licensing, please check this page.
This tool is helpful for fetching additional scripts/database files/directories from the Rosetta GitHub repository.
For example, if one's local machine does not have Rosetta built and installed, and wishes to check some files from $ROSETTA3_DB or use some helper scripts at $ROSETTA_PYTHON_SCRIPTS before run Rosetta tasks within Rosetta Container, one can use this tool to fetch them into the local harddrive by doing a minimum cloning.
The partial_clone function do will do the following steps:
- Check if Git is installed and versioned with
>=2.34.1. If not satisfied, raise an error to notify the user to upgrade git. - Check if the target directory is empty or not and the repository is not cloned yet.
- Setup partial clone and sparse checkout stuffs.
- Clone the repository and subdirectory to the target directory.
- Setup the environment variable with the target directory.
import os
from RosettaPy.utils import partial_clone
def clone_db_relax_script():
"""
A example for cloning the relax scripts from the Rosetta database.
This function uses the `partial_clone` function to clone specific relax scripts from the RosettaCommons GitHub repository.
It sets an environment variable to specify the location of the cloned subdirectory and prints the value of the environment variable after cloning.
"""
# Clone the relax scripts from the Rosetta repository to a specified directory
partial_clone(
repo_url="https://github.com/RosettaCommons/rosetta",
target_dir="rosetta_db_clone_relax_script",
subdirectory_as_env="database",
subdirectory_to_clone="database/sampling/relax_scripts",
env_variable="ROSETTA3_DB",
)
# Print the value of the environment variable after cloning
print(f'ROSETTA3_DB={os.environ.get("ROSETTA3_DB")}')Thanks for Windows Subsystem for Linux(WSL), we provide two simple ways to run Rosetta on Windows.
One must enable Windows Subsystem for Linux, then switch to WSL2 following the instructions on this page.
-
Install
Docker Desktopand enableWSL2 docker engine. -
Search for the Image
rosettacommons/rosetta:<label>where<label>is the version of Rosetta build one want to use.- Note: network proxies or docker registry mirror setting may be required for users behind the GFW.
-
Use
RosettaContainerclass as the run node, with the image name one just pulled. -
Make sure all the input files are using
LFending instead ofCRLF. This is fatal for Rosetta to parse input files.- Note: this issue now can be done by using a context manager
convert_crlf_to_lffromRosettaPy.utils.tools. Example:
from RosettaPy.utils.tools import convert_crlf_to_lf with convert_crlf_to_lf(input_file) as output_file: """Use `output_file` to replace `input_file`."""
- Note: this issue now can be done by using a context manager
-
Build Rosetta workflow with
RosettaPyand run it.
- Install any recent release of Linux Distribution (e.g.,
Ubuntu-22.04) and setup for the account. - Install build essential tools:
apt-get update && apt-get install build-essential git -y- Note: network proxies or apt repostory mirror setting may be required for users behind the GFW.
- Install MPI:
apt-get install mpich -y. MPICH is sufficient for Rosetta. - Install Python:
apt-get install python-is-python3 -y - Fetch the source code of Rosetta and un-tar it to anywhere convenient. e.g.
/opt/rosetta - Go to the source code directory and build it according to the Official Rosetta Build Documentation.
- Environment variables required by RosettaPy are:
ROSETTA_BIN: path to the Rosetta executablesROSETTA3_DB: path to the Rosetta databaseROSETTA_PYTHON_SCRIPTS: path to the Rosetta scripts
- Use
WslWrapperclass as the run node. Parameters:rosetta_bin:RosettaBinarywith in-wsldirname(the absolute path of Rosetta binary directory in WSL distro).distro: the name of the Linux Distribution. for example,'Ubuntu-22.04'user: the name one just setup as the user in the Linux Distribution.nproc: number of CPU cores to be used.prohibit_mpi: whether to prohibit MPI.
The RosettaFinder searches the following directories by default:
PATH, which is commonly used in dockerized Rosetta image.- The path specified in the
ROSETTA_BINenvironment variable. ROSETTA3/binROSETTA/main/source/bin/- A custom search path provided during initialization.
The Rosetta binaries are expected to follow this naming pattern:
rosetta_scripts[[.mode].oscompilerrelease]
- Binary Name:
rosetta_scripts(default) or specified. - Mode (optional):
default,mpi, orstatic. - OS (optional):
linuxormacos. - Compiler (optional):
gccorclang. - Release (optional):
releaseordebug.
For reg expression to match the basenames:
^(?P<binary_name>[\w]+)((\.(?P<mode>static|mpi|default|cxx11threadserialization|cxx11threadmpiserialization))?(\.(?P<os>linux|macos)(?P<compiler>gcc|clang)(?P<release>release|debug)))?$See this regex101 page for more details.
Examples of valid binary filenames:
rosetta_scripts(dockerized Rosetta)rosetta_scripts.linuxgccreleaserosetta_scripts.mpi.macosclangdebugrosetta_scripts.static.linuxgccrelease
The project includes unit tests using Python's pytest framework.
-
Clone the repository (if not already done):
git clone https://github.com/YaoYinYing/RosettaPy.git
-
Navigate to the project directory and install the required dependencies:
cd RosettaPy pip install '.[test]'
-
Run the tests:
# quick test cases pytest ./tests -m 'not integration' # test integration cases pytest ./tests -m 'integration' # run integration tests with both docker and local export GITHUB_CONTAINER_ROSETTA_TEST=YES pytest ./tests -m 'integration'
Contributions are welcome! Please submit a pull request or open an issue for bug reports and feature requests.
- Rosetta Commons: The Rosetta software suite for the computational modeling and analysis of protein structures.
- CIs, formatters, checkers and hooks that save my life and make this tool improved.
- ChatGPT, Tongyi Lingma, DeepSource Autofix™ AI and CodeRabbit for the documentation, code improvements, test cases generations and code revisions.
For questions or support, please contact:
- Name: Yinying Yao
- Email:yaoyy.hi(a)gmail.com
Footnotes
-
For building Rosetta upon aarch64, please refer to this example. ↩ ↩2 ↩3
-
Translated with Rosetta2 framework if runs on Apple Silicon Mac, which may cause worthy slow performance. ↩
-
Windows Subsystem for Linux(WSL) installed and switched to WSL2, with Rosetta built and installed on. ↩
-
It's theoretically possible yet no testing is done at all. ↩